Animal Applied Microbiology
Animal Applied Microbiology
Staff
 | Prof. Dr. MORITA Hidetoshi E-mail: hidetoshi-morita@(@okayama-u.ac.jp) Specialties: Food Function, Microbial Genomics Studies on human microbiome and comparative genomics of bacteria |
 | Assoc. Prof. Dr. ARAKAWA Kensuke E-mail: karakawa@(@okayama-u.ac.jp) Specialties: Dairy Chemistry, Food Microbiology Processing and quality control of animal food products using lactic acid bacteria |
Research Topics
1. The food composition and compornent drastically change human microbiota. Furthermore, recent insights on the link between the intestinal microbiota and human health are provided and changes in composition and diversity of human intestinal microbiota are related to disease. In our laboratory, we analyze the human intestinal gut and oral microbiota (human microbiome) and the genomics of human origine bacteria, bifidobacteria and lactic acid bacteria.
2. From ancient times, lactic acid bacteria (LAB) are widely used as microbial starter cultures for fermented foods to improve the taste and flavor, change the texture, and extend the shelf life. In the last several decades, it has been revealed that LAB have many beneficial effects not only for food processing and preservation but also for health maintenance and promotion (by working as probiotics and/or biogenics). In our laboratory, isolation, characterization and food application, particularly for milk products, of beneficial LAB strains and the metabolites are performed.
Analysis of human intestinal and oral microbiota (human microbiome)
|
Human and animal gut/oral microbiota are analyzed by next generation sequencer and their functions are studied by statistics (dry) and wet lab experiments. In particular, we research about the key bacteria in patient or excellent athlete microbiota by the comparison of healthy microbiota. In these researches, germ-free/gnotobiotic mice are powerful tools. | 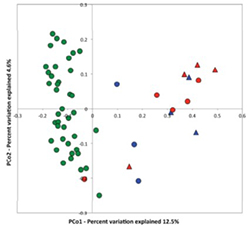 |
Genomics of Human origine bacteria, bifidobacteria and lactic acid bacteria
|
In general, the properties and functions of bacteria are different as not the species but the strains. The genome analysis of bacteria is the base of the various researches including their functional analysis. We also promote to contribute to exhaustive gene expression profiling analysis and epigenetics. | 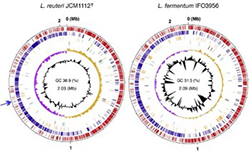 |
Studies on protein metabolism of lactic acid bacteria
Lactic acid bacteria (LAB) are known as fastidious microorganisms that require a wide variety of exogenous free amino acids or peptides as nitrogen sources for growth. Most of Food stuffs such as milk generally contain proteins but very low amount of free amino acids and peptides. Therefore, LAB first hydrolyze the proteins to get nutritious oligopeptides using their own cell-envelope proteinases (CEP). The oligopeptides released are subsequently taken up by the LAB cells via specific transport systems for further degradation into shorter peptides and free amino acids using various intracellular peptidases. The CEP, transporters and peptidases are species- or strain-specific, and their functional and enzymatic differences are an important factor to produce the diversity on taste, flavor and health-promoting effect of fermented food products. In our laboratory, we are studying the protein metabolism of LAB; particularly focusing on characterization of CEP and function of the oligopeptides released by CEP on fermented foods.
Studies on antimicrobial substances produced by lactic acid bacteria
Lactic acid bacteria (LAB) are known as safe producers of antimicrobial substances such as lactic acid and bacteriocins. Bacteriocins, ribosomally biosynthesized antibacterial peptides, have strong activity with a very small quantity and are easily degraded by digestive enzymes in the animal gut. Food-derived antimicrobial peptides are also produced by LAB. Food proteins such as milk caseins are good origins of functional peptides, including with antimicrobial activity, released by hydrolysis using cell-envelope proteinases (CEP) of LAB. These antimicrobial peptides are expected to be used as a safe natural preservative (biopreservative) for foods and feeds. In our laboratory, we isolated some LAB strains with bacteriocin productivity and high CEP activity for production of food-derived antimicrobial peptides, and characterized the bacteriocins and the food-derived antimicrobial peptides for application to foods and feeds.
Publication List
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W,Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S,Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nuñez G, Camp JG, Hattori M, Umesaki Y, Honda K. 2015. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 163, 367-380.
- Arakawa K, Matsunaga K, Takihiro S, Moritoki A, Ryuto S, Kawai Y, Masuda T, Miyamoto T. 2015. Lactobacillus gasseri requires peptides, not proteins or free amino acids, for the growth in milk. J Dairy Sci 98, 1593-1603.
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232-236.
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. 2013. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97-101.
- Toh H, Oshima K, Nakano A, Takahata M, Murakami M, Takaki T, Nishiyama H, Igimi S, Hattori M, Morita H. 2013. Genomic adaptation of the Lactobacillus casei group, PLoS ONE 8, e75073.
- Morita H, Nakano A, Onoda H, Toh H, Oshima K, Takami H, Murakami M, Fukuda S, Takizawa T, Kuwahara T, Ohno H, Tanabe S, Hattori M. 2011. Bifidobacterium kashiwanohense sp. nov., isolated from healthy infant faeces. Int J Syst Evol Microbiol 61, 2610-2615.
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543-549.