Plant Genome Dynamics Analiysis
•Screening of the active retrotransposon families
•DNA genotyping based on retrotransposon insertion sites by NGS
•Linkage map construction based on retrotransposon insertion polymorphism
Staff
 | Assoc. Prof. Yuki MONDEN E-mail: y_monden (please add @okayama-u.ac.jp) Specialty: Plant Breeding and Genetics |
Research Topics
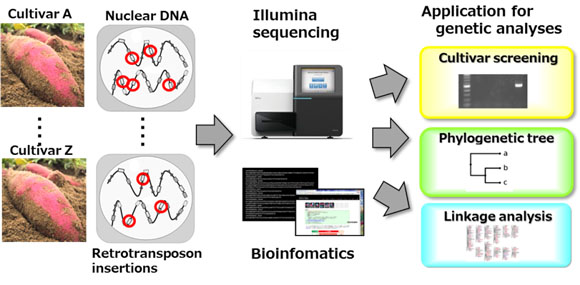
Retrotransposons are genetic elements that can replicate themselves through reverse transcription of their transcribed RNA and integration of the resulting cDNAs into new genomic loci in a genome. In higher plant genomes, retrotransposons are a major component of nuclear DNA. As retrotransposon insertions with high copy numbers are dispersed throughout the genome and are inherited genetically, different insertion sites among crop cultivars have been used as molecular markers. In our laboratory, a series of Next-Generation Sequencer (NGS) applications are developed to utilize retrotransposons as distinctive genetic analysis systems for several crops species.
Keyword: retrotransposon, NGS (next-generation sequencing), polyploidy, sweet potato (Ipomoea batatas (L.) Lam), molecular marker, DNA polymorphism, linkage analysis
Screening of the active retrotransposon families
As described above, different retrotransposon insertion sites among crop cultivars are useful in development of molecular markers. However, identification of the active families is challenging as they are concealed by numerous non-active families. Thus, we conducted an efficient screening of the retrotransposon families that showed high insertion polymorphism among closely related cultivars using NGS platform.
DNA genotyping based on retrotransposon insertion sites by NGS
Genotyping based on the polymorphic retrotransposon insertions has been conducted, which can be applied in phylogenetic, genetic diversity studies and the development of cultivar-screening markers. We identified the retrotransposon insertion sites at the genome-wide scale in a number of cultivars with a NGS platform. Our results indicated that the target sequencing of active retrotransposon insertion sites is highly effective for DNA genotyping without requiring any whole genome sequence information. In addition, the cultivar-specific insertion sites are used to develop a practical cultivar identification marker for not only direct products of a crop but also cultivar-blended processed products.
Linkage map construction based on retrotransposon insertion polymorphism
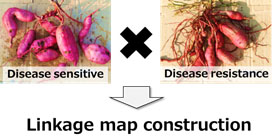
We have constructed a linkage map using retrotransposon insertion sites to identify genes of agronomic importance. The linkage analyses were conducted using the mapping populations derived from a cross between two different cultivars. We found that these insertion sites were quite useful for linkage map construction in polyploid species, as most insertion sites segregated at a simple ratio (1:1) in hexaploid sweet potato (2n=6x=90) mapping populations. Thus, it is expected that identifying a number of retrotransposon insertions will facilitate linkage map construction in polyploid species for quantitative trait loci (QTL) mapping and marker-assisted selection for plant breeding.
Please feel free to contact us if you have some interests of our research!
Publications
• Yuki Monden, Takuya hara, Yoshihiro Okada, Osamu Jahana, Akira Kobayashi, Hiroaki Tabuchi, Shoko Onaga, Makoto Tahara. Construction of a linkage map based on retrotransposon insertion polymorphisms in sweet potato via high-throughput sequencing. Breeding Science, accepted.
• Makoto Tahara, Yuki Monden, Erika Saiki. Efficient identification of retrotransposon families showing different insertion profile among soybean cultivars by the next-generation sequencer. DNA Polymorphism (DNA Takei), accepted. (In Japanese)
• Yuki Monden, Kentaro Yamaguchi, Makoto Tahara. Application of iPBS in high-throughput sequencing for the development of retrotransposon-based molecular markers. Current Plant Biology, 1, 40-44 (2014).
• Yuki Monden, Nobuyuki Fujii, Kentaro Yamaguchi, Kazuho Ikeo, Yoshiko Nakazawa, Takamitsu Waki, Keita Hirashima, Yosuke Uchimura, Makoto Tahara. Efficient screening of long terminal repeat retrotransposons that show high insertion polymorphism via high-throughput sequencing of the PBS site. Genome, 57(5): 245-252 (2014).
• Yuki Monden, Ayaka Yamamoto, Akiko Shindo, Makoto Tahara. Efficient DNA fingerprinting based on the targeted sequencing of active retrotransposon insertion sites using a bench-top high-throughput sequencing platform. DNA research, 21(5): 491-498 (2014).
• Yuki Monden, Kazuto Takasaki, Satoshi Futo, Kousuke Niwa, Mitsuo Kawase, Hiroto Akitake, Makoto Tahara. A rapid and enhanced DNA detection method for crop cultivar discrimination. Journal of Biotechnology, 185: 57-62 (2014).
• Yuki Monden, Takeru Takai, Makoto Tahara. Characterization of a novel retrotransposon TriRe-1 using nullisomic-tetrasomic lines of hexaploid wheat. The Scientific Reports of the Faculty of Agriculture, 103: 21-30 (2014).
• Yuki Monden, Takeru Takai, Makoto Tahara, Yuta Umeno, Ryota Nakamura. High-throughput development of DNA markers for wheat cultivar discrimination based on an active retrotransposon TriRe-1 insertion polymorphism. DNA Polymorphism (DNA Takei), 22(1): 60-65 (2014). (In Japanese)
• Yuki Monden, Ayaka Yamamoto, Makoto Tahara. Development of DNA markers for anthocyanin content purple sweet potato using active retrotransposon insertion polymorphisms. DNA Polymorphism (DNA Takei), 21: 47–54 (2013). (In Japanese)
• Hiroto Akitake, Makoto Tahara, Yuki Monden, Kazuto Takasaki, Satoshi Futo. Strawberry cultivar identification by retrotransposon insertion polymorphisms. DNA Polymorphism (DNA Takei), 21: 64–72 (In Japanese).